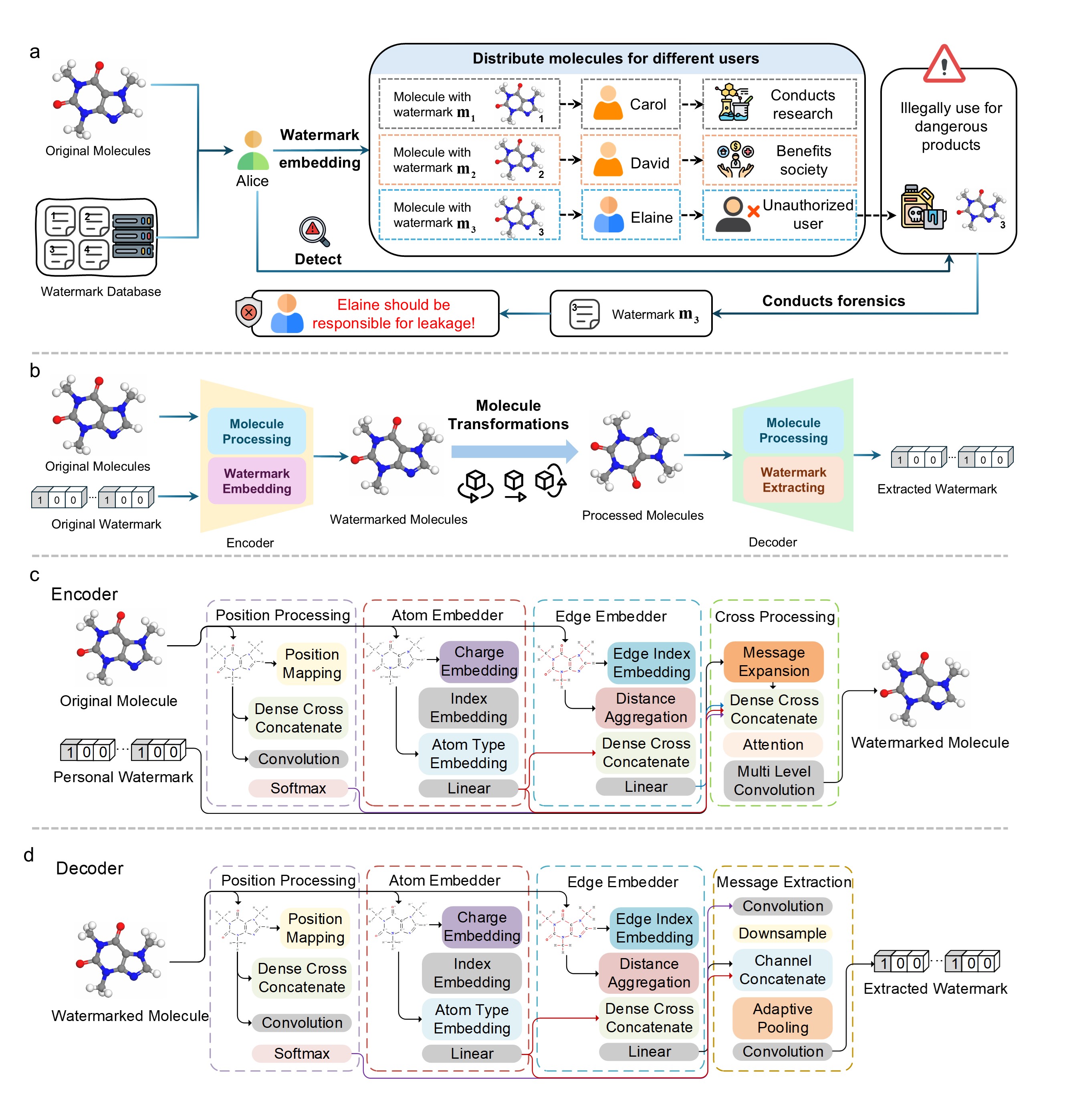
MolMark is the first dedicated watermarking framework for AI-generated molecular structures. MolMark operates at the atom level, embedding watermarks by subtly modulating chemically informed features, while strictly preserving molecular stability and function. Crucially, watermark encoding and extraction rely on SE(3)-invariant interatomic distance representations, ensuring robustness against geometric transformations.
Abstract
AI-driven molecular generation has revolutionized drug discovery and materials science, yet a critical gap remains: no tools exist to safeguard these AI-generated assets from intellectual property (IP) vulnerabilities, such as unauthorized reuse or ambiguous provenance. Here we present MolMark, the first dedicated watermarking framework tailored for molecular structures, engineered to embed robust digital signatures while preserving molecular function. MolMark leverages atom-level features (atomic type, charge, and interatomic constraints) to minimize structural perturbation during embedding, and extracts SE(3)-invariant interatomic distance features to ensure robustness against rotation, translation, and reflection. Validated on benchmark datasets (QM9, GEOM-DRUG) and state-of-the-art molecular generative models (GeoBFN, GeoLDM), 16-bit watermarks retained >90% of key molecular properties, achieved >95% extraction accuracy post-SE(3) transformations, and maintained near-identical performance in downstream molecular docking. MolMark bridges IP protection and scientific utility, enabling ethical AI-driven molecular discovery.
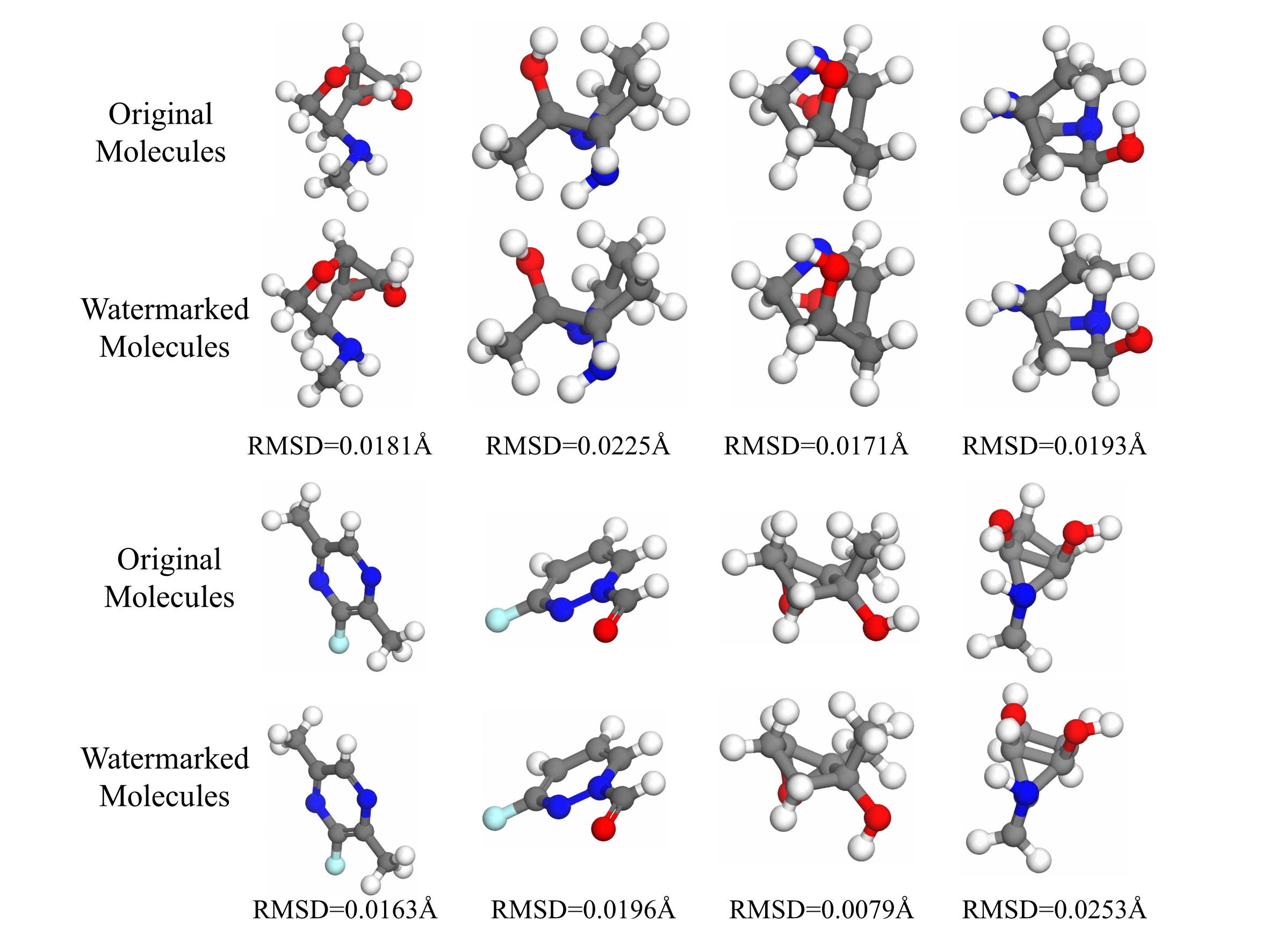
Structure of eight pairs of molecules. The original molecule and watermarked molecules are arranged vertically, in which the structures only undergo slight changes after embedding watermarks.
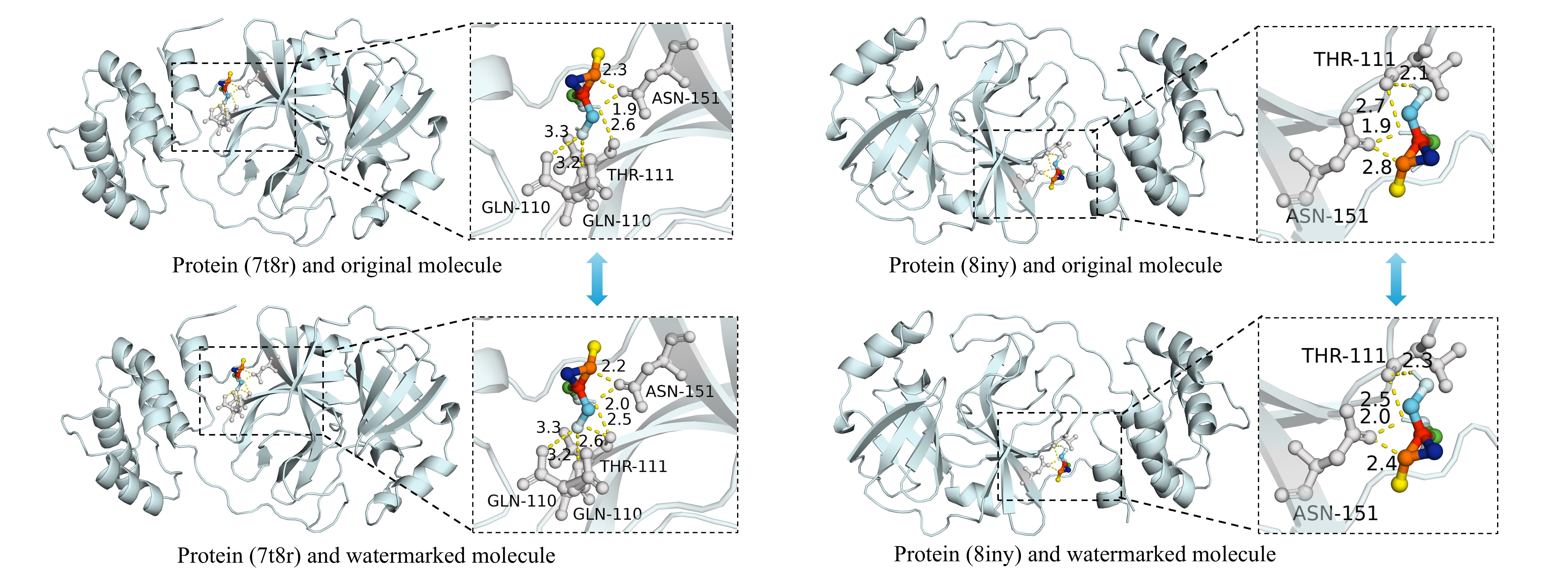
The docking rendering of original molecules and watermarked molecules with proteins. The amino acid residues and hydrogen bond lengths in molecular docking are similar, reflecting that MolMark has an insignificant impact on the effectiveness of molecules.
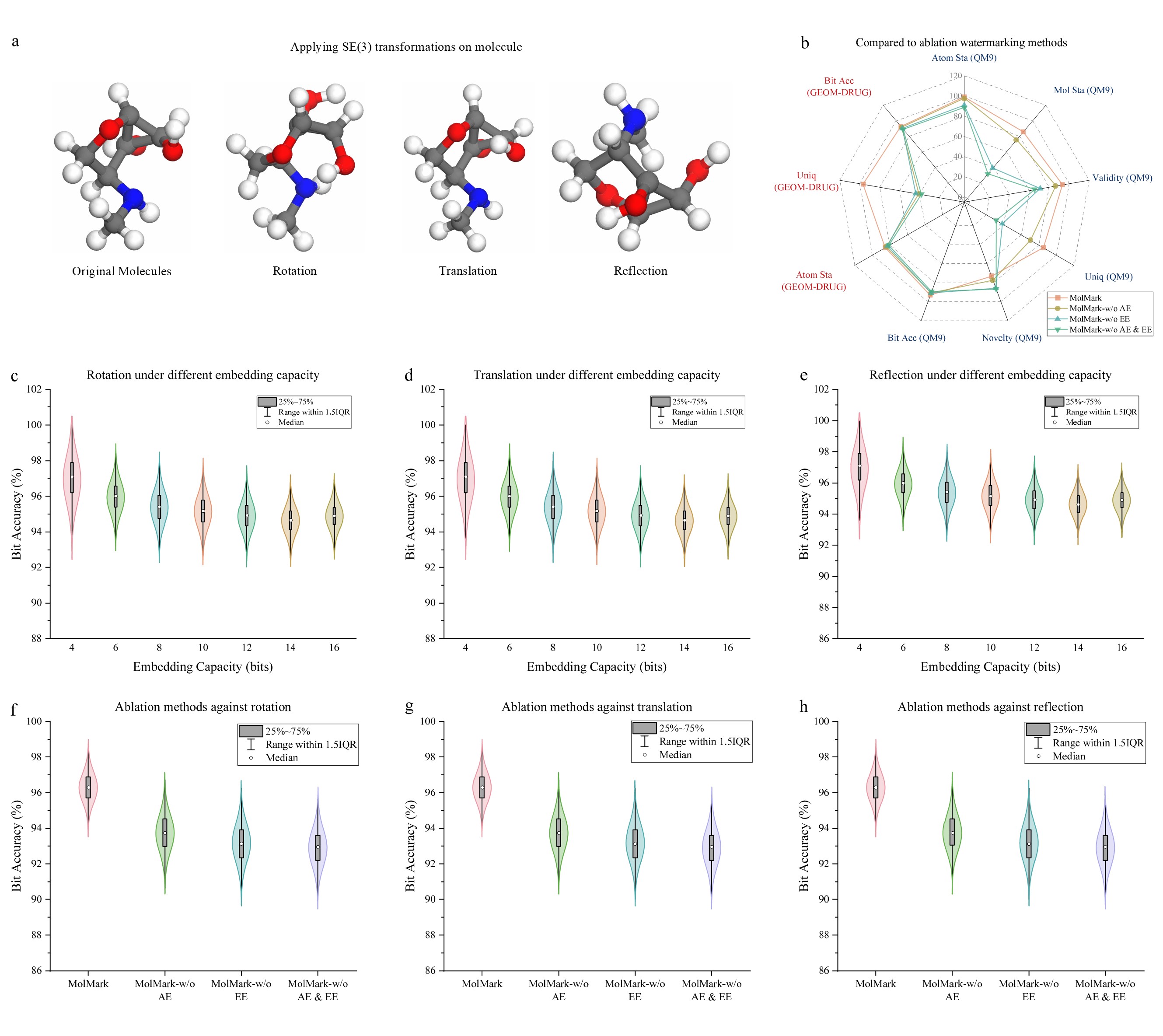
Robustness against SE(3) transformations. The confirmations of molecule after applying SE(3) transformations, including rotation, translation, and reflection. The conformations of the processed molecules are different, but they share the same characteristics. The bit accuracies under SE(3) transformations are listed as follows.
BibTeX
@article{hu2024molmark,
title={MolMark: Safeguarding Molecular Structures with Atom-Level Watermarking},
author={Hu, Runen and Chen, Peilin and Ding, Keyan and Wang, Shiqi},
journal={arXiv},
year={2025},
}